REST API / Web UI for similarity searches
This is an example of using the supplied command line tools to generate descriptors for molecule sets and start up an embedded server to provide a Web UI and REST API for remote clients. Parts of the steps described below are implemented in script rest-api-example.sh found in examples/ directory. Further scripts exposing larger datasets and more descriptors are available. For details see document Self contained examples. This documentation also details core concepts of command line tool gui.sh.
Basic workflow consists of the following steps:
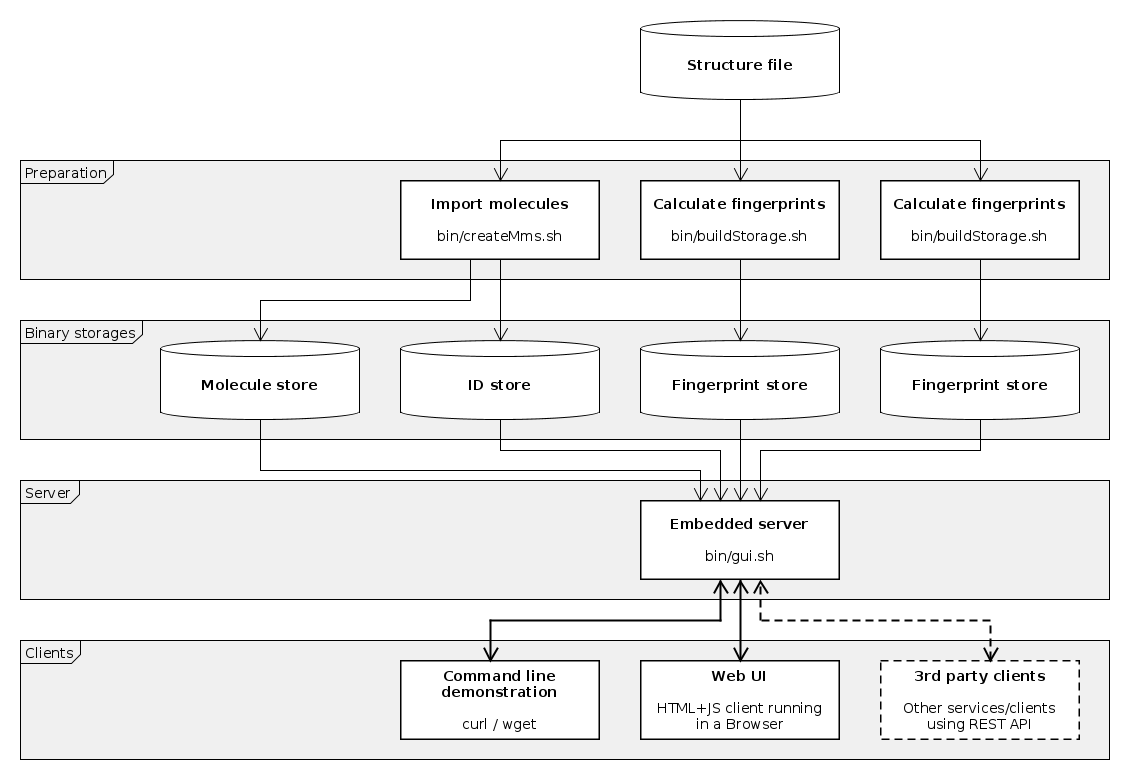
- Import molecules and ID from structure file (creating master molecule storage and master ID storages).
- Calculate molecular descriptors to be used.
- Launch embedded server to provide REST API and serve Web UI for clients.
- Connect using tool
curl(orwget) to query from bash command line. - Connect using a browser to provide an interactive user interface (Web UI).
For more details on the command line scripts involved see their description. See also introduction to REST API slides.
Process and expose contents of data/vitamins.smi
Expose the vitamins dataset containing 30 structures with a simple descriptor (CFP7) and a set of custom float descriptors.
Commands
# Import molecules and IDs
cat data/molecules/vitamins/vitamins.smi | bin/createMms.sh \
-in - \
-name vitamins-name.bin -out vitamins-mms.bin
# Calculate CFP7 descriptors
cat data/molecules/vitamins/vitamins.smi | bin/buildStorage.sh \
-context createSimpleCfp7Context \
-in - \
-out vitamins-cfp7.bin
# Import custom float descriptors
cat data/floats-1d.txt | bin/importStorage.sh \
-in - \
-splitter com.chemaxon.overlap.splits.AllButFirstToken \
-idsplitter com.chemaxon.overlap.splits.FirstToken \
-out custom-float-1d-desc.bin \
-id custom-float-1d-id.bin \
-contextjs "ctx_from_descpb(bld_fv.length(1))" \
-infilter "(l.trim().length == 0 || l.trim().charAt(0) == '#') ? null : l"
# Launch embedded server
bin/gui.sh \
-mols -name:vitamins:-mms:vitamins-mms.bin:-mid:vitamins-name.bin \
-idonly -name:custom-1d-float:-mid:custom-float-1d-id.bin \
-desc -desc:vitamins-cfp7.bin:-mols:vitamins:-name:vita-cfp7 \
-desc -desc:custom-float-1d-desc.bin:-mols:custom-1d-float:-name:custom-1d-float \
-nobrowse \
-port 8085 \
-stopport 8086 \
-stopsecret my_stop_secret
After startup messages similar for the following example are printed to the console:
....
Server stopper listening on port 8086. Open connection and send secret to stop server.
Server listening on port 8085
Try connect to http://localhost:8085/index.html
Or to the following network interfaces:
em1 (em1)
http://192.168.1.133:8085/index.html
lo (lo)
http://127.0.0.1:8085/index.html
Parametrization and usage of tools createMms.sh, buildStorage,sh are described in document Basic search workflow. Tool importStorage introduced in documents Using custom binary descriptors and Using custom float descriptors.
Details on parametrization of gui.sh:
When parameter -stopsecret (and optionally -stopport) specified the server can be stopped by connecting to the specified (or default) port and sending the specified secret. See example below.
When parameter -nobrowse is missing the tool tries to launch the default web browser pointing to the initial page exposed by the embedded server.
Parameter -port specifies the port on which the server listens. The REST API and the static contents of the Web UI both are served on this port. Value 0 can be passed to parameter -port. In this case an available port is chosen for listening. The number of the chosen port is printed to the console.
Parameter -mols specifies molecule storage (exposed under REST resource rest/molecules/<NAME>/). Argument for this parameter is a : separated list of further arguments specifying the details of the molecule storage:
| Sub-parameter name | Value in this example | Description |
|---|---|---|
-name |
vitamins |
<NAME> be to used when exposed as resource rest/molecules/<NAME>/ |
-mms |
vitamins-mms.bin |
File to read master molecule storage containing structures |
-mid |
vitamins-name.bin |
File containing molecule IDs for the specified master molecule storage |
Parameter -idonly specifies a molecule storage storing only IDs. All the associated molecules are marked as absent. This allows the association of IDs to custom descriptors without attached structure sources.
| Sub-parameter name | Value in this example | Description |
|---|---|---|
-name |
custom-1d-float |
<NAME> be to used when exposed as resource rest/molecules/<NAME>/ |
-mid |
custom-float-1d-id.bin |
File containing IDs to be exposed |
Parameter -desc specifies descriptors (exposed under REST resource rest/descriptors/<NAME>/). Argument for this parameter is a : separated list of further arguments specifying the details of the descriptor storage:
| Sub-parameter name | Value in this example | Description |
|---|---|---|
-name |
vita-cfp7 |
<NAME> be to used when exposed as resource rest/descriptors/<NAME>/ |
-desc |
vitamins-cfp7.bin |
File to read descriptor storage from |
-mols |
vitamins |
<NAME> of associated -mols resource containing the molecules |
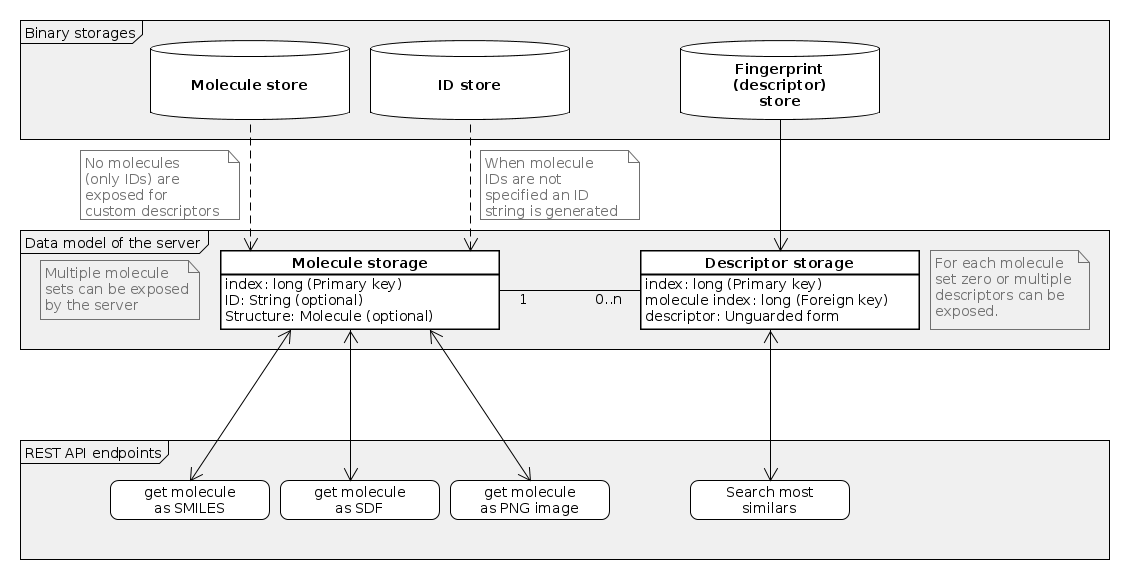
Structure of the exposed data
The exposed data (molecules, IDs and searchable descriptors) can be viewed as a simple relational data model.
Using Web UI
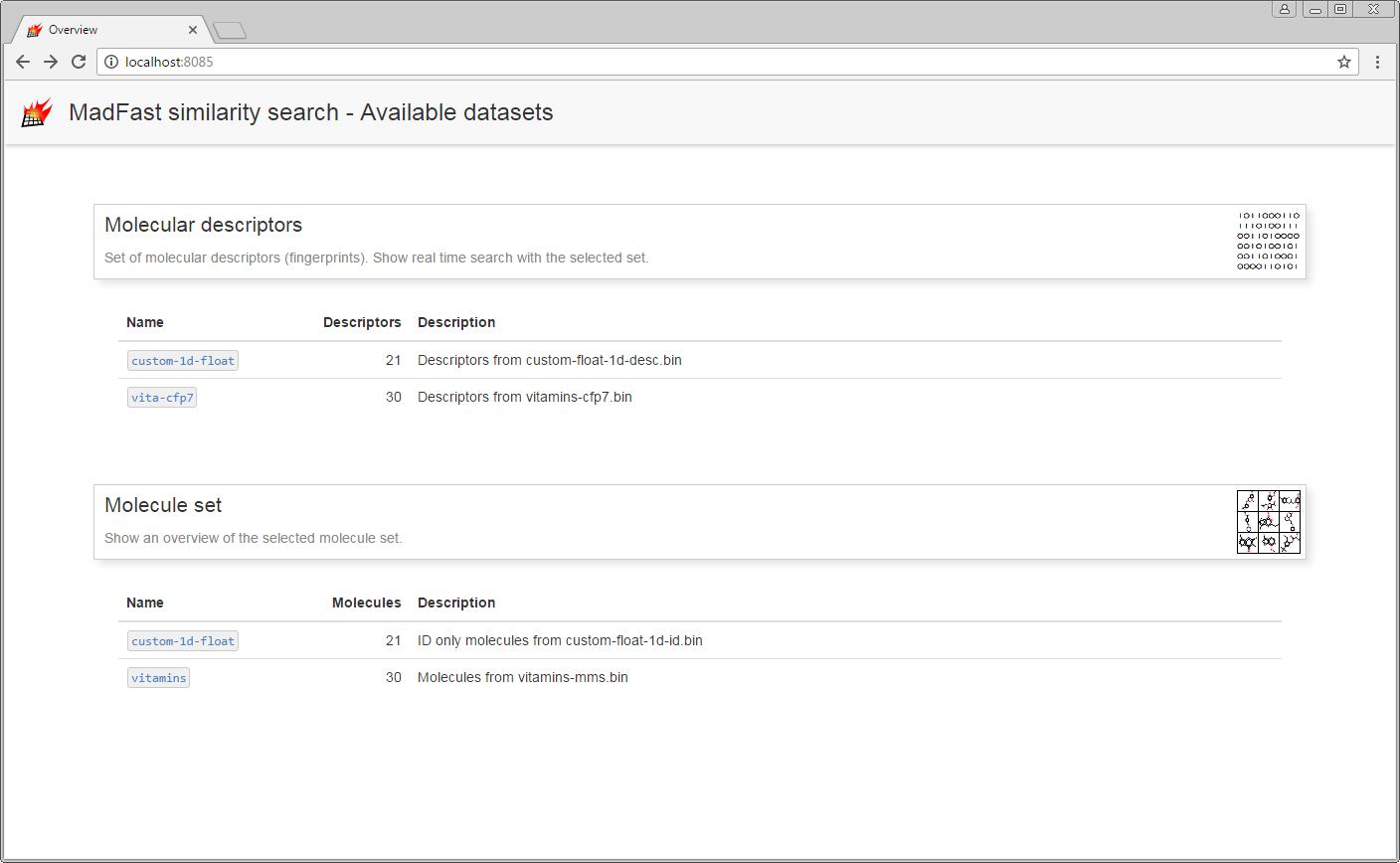
After launching the server connect from a browser to http://localhost:8085. The overview page with the available resources (data served/searchable by the server) is presented. In the current example two molecule sets and one associated descriptor for both are available. The vitamins molecule set can be browsed; the descriptor associated to it (vita-cfp) can be queried using structure queries. The imported custom float descriptors (custom-1d-float) and its associated virtual molecule storage containing the imported IDs (custom-1d-float) currently can not be queried or handled by the web UI.
Connecting to the embedded server from command line
Query available molecular descriptors (parameter -g passed to curl switched off "URL globbing parser" so URLs containing letters {}[] can be specified):
curl -g "http://localhost:8085/rest/descriptors"
Alternatively wget can be used (parameter -qO0 passed to wget turns off wget output and writes downloaded content to stdout):
wget -qO- "http://localhost:8085/rest/descriptors"
Output is a JSON object describing the available descriptors which can be used as a search target:
{"descriptors":[{"size":30,"description":"Descriptors from vitamins-cfp7.bin","context":"Overlap analysis context.\n Pagesize: 50\n Standardizer: ThreadLocalized wrapper over chemaxon.standardizer.Standardizer@d255df2 (actions count: 1)\n Generator: CFP generator, parameters: bond count: 7 (bits per pattern: 1, length: 1024)\n Comparator: Comparator BINARY_TANIMOTO, vector size: 1024 bits\n Extractor: Extract packed long [] fingerprint representation (16 longs, 1024 bits)\n Unguarded calc: Tanimoto dissimilarity of binary fingerprints represented as packed long[]\n","moleculeseturl":"rest/molecules/vitamins","url":"rest/descriptors/vita-cfp7","name":"vita-cfp7"}]}
If Python 2.6+ available the output can be formatted:
curl -g "http://localhost:8085/rest/descriptors" | python -m json.tool
{
"descriptors": [
{
"context": "Overlap analysis context.\n Pagesize: 50\n Standardizer: ThreadLocalized wrapper over chemaxon.standardizer.Standardizer@d255df2 (actions count: 1)\n Generator: CFP generator, parameters: bond count: 7 (bits per pattern: 1, length: 1024)\n Comparator: Comparator BINARY_TANIMOTO, vector size: 1024 bits\n Extractor: Extract packed long [] fingerprint representation (16 longs, 1024 bits)\n Unguarded calc: Tanimoto dissimilarity of binary fingerprints represented as packed long[]\n",
"description": "Descriptors from vitamins-cfp7.bin",
"moleculeseturl": "rest/molecules/vitamins",
"name": "vita-cfp7",
"size": 30,
"url": "rest/descriptors/vita-cfp7"
}
]
}
Invoke similarity searches
Both GET and POST requests are supported. Invoke a GET request using URL encoded query parameters:
curl -g "http://localhost:8085/rest/descriptors/vita-cfp7/find-most-similars?count=4&query=C([C@@H]([C@@H]1C(=C(C(=O)O1)O)O)O)O" | \
python -m json.tool
Invoke a POST request:
curl \
-X POST \
-d "count=4" \
-d "query=C([C@@H]([C@@H]1C(=C(C(=O)O1)O)O)O)O" \
-g \
"http://localhost:8085/rest/descriptors/vita-cfp7/find-most-similars" | python -m json.tool
The results for both requests are the same:
{
"query": "C([C@@H]([C@@H]1C(=C(C(=O)O1)O)O)O)O",
"querysmi": "C([C@@H]([C@@H]1C(=C(C(=O)O1)O)O)O)O",
"searchtime": 2,
"targets": [
{
"base64img": null,
"dissimilarity": 0.0,
"targetid": "Vitamin C - Ascorbic acid",
"targetimageurl": "rest/molecules/vitamins/16/png?w=100&h=100",
"targetindex": 16,
"targetmolurl": "rest/molecules/vitamins/16"
},
{
"base64img": null,
"dissimilarity": 0.77450980392156865,
"targetid": "Vitamin D3 - Cholecalciferol",
"targetimageurl": "rest/molecules/vitamins/17/png?w=100&h=100",
"targetindex": 17,
"targetmolurl": "rest/molecules/vitamins/17"
},
{
"base64img": null,
"dissimilarity": 0.78301886792452835,
"targetid": "Vitamin D3 - Ergocalciferol",
"targetimageurl": "rest/molecules/vitamins/18/png?w=100&h=100",
"targetindex": 18,
"targetmolurl": "rest/molecules/vitamins/18"
},
{
"base64img": null,
"dissimilarity": 0.81188118811881194,
"targetid": "Vitamin A - Retinol",
"targetimageurl": "rest/molecules/vitamins/0/png?w=100&h=100",
"targetindex": 0,
"targetmolurl": "rest/molecules/vitamins/0"
}
]
}
Access targets
Reference targetmolurl and targetimageurl can be used to access targets:
curl "http://localhost:8085/rest/molecules/vitamins/17/png?w=100&h=100" > hit.png
curl "http://localhost:8085/rest/molecules/vitamins/17/id"
curl "http://localhost:8085/rest/molecules/vitamins/17/smiles"
Ids for multiple targets can be queried in a single batch:
curl -X POST \
-H "Content-Type: application/x-www-form-urlencoded" \
-d 'indices[]=10&indices[]=11&indices[]=12' \
-g "http://localhost:8085/rest/molecules/vitamins/get-multiple-ids" | python -m json.tool
Closing the server
If option -stopsecret is specified the server can be stopped by opening a TCP connection to the port specified by option -stopport and sending the specified secret. One can use tool netcat on linux:
echo "my_stop_secret" | nc localhost 8086
Notes on URL encoding
Query SMILES parameter in the query string must be URL encoded. One possible tool available as part of standard Java SE distributions is java.net.URLEncoder.encode(String s, String encoding).
This method can be invoked from command line tool jseval through the provided scripting hook:
bin/jseval.sh -d "string=Special characters: & ? [ ] #" -js "println('ENCODED: ' + java.net.URLEncoder.encode(string, 'UTF-8'));"
com.chemaxon.overlap.cli.JsEval
args: [-d, string=Special characters: & ? [ ] #, -js, println('ENCODED: ' + java.net.URLEncoder.encode(string, 'UTF-8'));]
Use parameter name: "string" value: "Special characters: & ? [ ] #"
JavaScript code to be executed:
println('ENCODED: ' + java.net.URLEncoder.encode(string, 'UTF-8'));
Launch.
ENCODED: Special+characters%3A+%26+%3F+%5B+%5D+%23
(Finished) Execution time: 21 ms, no invocations
All done.
Please note that command println was used in the scripting hook. Support for println varies with the script engine shipped with the Java runtime. Tool jseval uses the workaround suggested in https://bugs.openjdk.java.net/browse/JDK-8035181 to provide println support for the Nashorn script engine shipped with jdk8.
Details on the parametrization of jseval used:
| Parameter name | Value in this example | Description |
|---|---|---|
-d |
string=Special characters: & ? [ ] # |
Parameter name and value to expose in the javascript execution context. The exposed parameter name is the part of the value before character -. |
-js |
println('ENCODED: ' + java.net.URLEncoder.encode(string, 'UTF-8')); |
JavaScript code to execute. Note that value for string is specified by parameter -d passed to jseval. |
Notes on error handling
REST API endpoints return a status descriptor in JSON format in case of an error. See diagnostic API endpoint /rest/generate-error-response for details (endpoint documentation).
Advanced server configuration: Use SSL (https)
Options -sslkeystore and -sslkeystorepass can specify an SSL keystore. If specified the embedded server will listen for https connections.
To create a self signed certificate with keytool (part of Java distributions; see its documentation). WARNING! This certificate is generated for demonstration, do not use it in a production environment.
Generate self signed certificate
keytool \
-genkey -noprompt -keyalg RSA -alias "my-alias" -validity 365 -keystore my-keystore.jks -keysize 2048 \
-storepass "32d0cca92adca483650da9778efb8aa1c" \
-keypass "32d0cca92adca483650da9778efb8aa1c" \
-dname "cn=cn value, ou=ou value, o=o value, c=cc"
Init and launch server
# Import molecules and IDs
cat data/molecules/vitamins/vitamins.smi | bin/createMms.sh \
-in - \
-name vitamins-name.bin -out vitamins-mms.bin
# Calculate CFP7 descriptors
cat data/molecules/vitamins/vitamins.smi | bin/buildStorage.sh \
-context createSimpleCfp7Context \
-in - \
-out vitamins-cfp7.bin
# Import custom float descriptors
cat data/floats-1d.txt | bin/importStorage.sh \
-in - \
-splitter com.chemaxon.overlap.splits.AllButFirstToken \
-idsplitter com.chemaxon.overlap.splits.FirstToken \
-out custom-float-1d-desc.bin \
-id custom-float-1d-id.bin \
-contextjs "ctx_from_descpb(bld_fv.length(1))" \
-infilter "(l.trim().length == 0 || l.trim().charAt(0) == '#') ? null : l"
# Launch embedded server
bin/gui.sh \
-mols -name:vitamins:-mms:vitamins-mms.bin:-mid:vitamins-name.bin \
-idonly -name:custom-1d-float:-mid:custom-float-1d-id.bin \
-desc -desc:vitamins-cfp7.bin:-mols:vitamins:-name:vita-cfp7 \
-desc -desc:custom-float-1d-desc.bin:-mols:custom-1d-float:-name:custom-1d-float \
-nobrowse \
-port 8085 \
-stopport 8086 \
-stopsecret my_stop_secret \
-sslkeystore my-keystore.jks \
-sslkeystorepass 32d0cca92adca483650da9778efb8aa1c
Please note that example script rest-api-example.sh does not demonstrate SSL configuration described here.
Connect
When launched connect with a browser to https://localhost:8085. Note that you have to manually add an exception to force browser to accept the self signed certificate. Alternatively curl can be used (since we are using a self signed certificate option -k needed for alllowing "insecure" connection):
curl -gk "https://localhost:8085/rest/descriptors/vita-cfp7" | python -m json.tool
{
"context": "Overlap analysis context.\n Pagesize: 50\n Standardizer: ThreadLocalized wrapper over chemaxon.standardizer.Standardizer@72dba68d (actions count: 1)\n Generator: CFP bond count: 7 (bits per pattern: 1, length: 1024)\n Comparator: Comparator BINARY_TANIMOTO, vector size: 1024 bits\n Extractor: Extract packed long [] fingerprint representation (16 longs, 1024 bits)\n Unguarded calc: Tanimoto dissimilarity of binary fingerprints represented as packed long[]\n",
"description": "Descriptors from vitamins-cfp7.bin",
"moleculeseturl": "rest/molecules/vitamins",
"name": "vita-cfp7",
"size": 30,
"url": "rest/descriptors/vita-cfp7"
}
Advanced server configuration: Use cross-origin resource sharing
To support Cross-Origin Resource Sharing use parameter -allowedOrigins <ORIGINS>. When this parameter is specified CrossOriginFilter is configured. The value of the parameter <ORIGINS> is used as the allowedOrigins parameter of the filter.
Please note that example script rest-api-example.sh demonstrates CORS configuration described here.
Demonstrate using curl
bin/gui.sh -nobrowse -allowedOrigins "*,*"
# from a different terminal while command above still running
curl -i -H "Origin: foo.bar" http://localhost:8081/rest/descriptors
HTTP/1.1 200 OK
Date: Tue, 25 Oct 2016 21:17:55 GMT
Access-Control-Allow-Origin: foo.bar
Access-Control-Allow-Credentials: true
Content-Type: application/json
Content-Length: 18
Server: Jetty(9.3.13.v20161014)
{"descriptors":[]}
Demonstrate using browsers
In the following example two servers are launched (in different terminals) to listen on different ports.
# Launch embedded server 1 - with no CORS
bin/gui.sh \
-mols -name:vitamins:-mms:vitamins-mms.bin:-mid:vitamins-name.bin \
-desc -desc:vitamins-cfp7.bin:-mols:vitamins:-name:vita-cfp7 \
-nobrowse \
-port 8085
# Launch embedded server 2 in a different terminal - with CORS
bin/gui.sh \
-mols -name:vitamins:-mms:vitamins-mms.bin:-mid:vitamins-name.bin \
-desc -desc:vitamins-cfp7.bin:-mols:vitamins:-name:vita-cfp7 \
-nobrowse \
-port 8086 \
-allowedOrigins "*,*"
Note that *,* used as the value of -allowedOrigins. This is a workaround for a problem with command line arguments globbing when when using Windows + Cygwin.
Both servers expose real time search for the vitamins datasets, all links (using absolute and relative references) work:
- http://localhost:8085/simsearch.html?ref=rest/descriptors/vita-cfp7/
- http://localhost:8085/simsearch.html?ref=http://localhost:8085/rest/descriptors/vita-cfp7/
- http://localhost:8086/simsearch.html?ref=rest/descriptors/vita-cfp7/
- http://localhost:8086/simsearch.html?ref=http://localhost:8086/rest/descriptors/vita-cfp7/
Page served by the CORS enabled server (listening on port 8086) can not fetch data from non CORS enabled (listening on port 8085) server, following link breaks:
Page served by non CORS enabled (listening on port 8085) server can fetch data from CORS enabled (listening on port 8086) server, following link works:
Advanced server configuration: Use request logging
Tool gui.sh can write a text based access log of the embedded server when using option -log <LOGFILE>. Please note that the log file format might be changed in the future releases and it does not contain the POST request bodies. Request log is written by org.eclipse.jetty.server.NCSARequestLog provided by the embedded Jetty server.
Advanced server configuration: Additional static content
Additional static content can be exposed by the embedded server gui.sh by option -additionalresourcedir <DIR>. When specified contents of the given directory <DIR> will be exposed under /additional/. MarvinJS used in the WebUI can recognize a Marvin JS license file (marvin4js-license.cxl) put into the given directory.
Note that an alternative mechanism is available to serve additional content, see document Raw file handling for details.
Advanced server configuration: Feature flags
Certain functionalities of the embedded server can be disabled by option -disable <FEATURE>. For a list of available features invoke gui.sh -h.
Some of these features are relevant for securely deploying the embedded server. Please see document REST API security considerations for details.