Getting started guide
Prerequisites
-
JAVA: This MadFast release requires Java version 8. According to ChemAxon's Supported Java Flavors and Versions we recommend to use AdoptOpenJDK. MadFast is expected to run other Java 8 distributions, including Oracle Java 8.
-
MEMORY: Due to the memory intensive nature of these tools 64 bit operation system and 64 bit java is recommended. Memory requirements are dependent on the number of structures and descriptors exposed. Using 1024 bit fingerprints and ususal small molecules the expected memory requirement is one GB per 3-4 million structures. See Server memory optimization for details.
-
LICENSE: License
MADFASTis required for the core functionality. LicenseECFPneeded for usingECFPfingerprint family, licenseMACCSis required forMACCS-166fingerprints. License forName to structureand forMarvin JSmight needed for certain functionalities.Further licenses might be required when additional properties are calculated through Chemical Terms (see Store additional data).
For further questions or to obtain an evaluation or production license feel free to contact us at
sales@chemaxon.comordisco-support@chemaxon.com.
Additional prerequisites on Windows
The current version is supplied with bash launcher scripts requiring linux or windows + cygwin to run. An illustrated guide for installing the prerequisites (including CygWin) on Windows is available in Installing dependencies on Windows version 10.
Additional prerequisites on Max OS X
Comand line scripts use greadlink instead of readlink on Max OS X internally. It can be installed using brew install coreutils. For details see https://brew.sh/.
Example script download-molecules.sh and some documentation examples use wget; it can be installed using brew install wget.
Install license file
Make sure that supplied license.cxl file is installed: the license file should be copyed to its default location. The default location is .chemaxon/license.cxl (on Unix) or chemaxon\license.cxl (on Windows + Cygwin). For details see ChemAxon Installing Licenses documentation.
Note that Marvin JS license stored in a separate file, see REST API / Web UI for similarity searches section Advanced server configuration: Additional static content and Self contained examples for details.
Install license file on Linux / Mac OS X
Evaluation / production license file license.cxl should be placed in directory .chemaxon in the home of the user running MadFast.
mkdir -p ~/.chemaxon/
cp license.cxl ~/.chemaxon/
Install license file on Windows
Evaluation / production license file license.cxl should be placed in directory chemaxon in the home of the user running MadFast. Typically the user home directory is c:\Users\<USERNAME>\. From a cygwin terminal
mkdir -p /cygdrive/c/Users/<USERNAME>/chemaxon
cp license.cxl /cygdrive/c/Users/<USERNAME>/chemaxon/
Unpack distribution
Unpack the contents of the downloaded archive. No further compilation or setup needed before invoking the command line interfaces or examples. Note that the launcher scripts in this version need to be invoked directly without using links pointing to them. Further examples will use paths relative to the distributions root directory.
tar xvf madfast-cli-0.3.5.tar
cd madfast-cli-0.3.5/
By default the self contained example scripts (detailed below) use the examples-tmp/ directory to write workfiles and results. Make sure that this directory is writable by the user. Please note that this is usually not required in production usage.
Verify
To verify launch an executable:
bin/createMms.sh -h
A help message similar to the following is expected to appear:
Usage: <main class> [options]
Options:
-h, -help, --help
Print help on usage then exit
Default: false
-count
Max number of structures to input
Default: 2147483647
....
Launch self contained example
Self contained example scripts found in directory examples/. For further details see their description.
Simple search workflow
Launch script examples/search-workflow.sh. After the preparation steps this script launches similarity searches against the drugbank dataset. For more details on the executed workflow see document Basic search workflow. The execution log file and the search results can be found in the work directory which default is examples-tmp/search-workflow/ in the distribution directory.
# launch self contained example
./examples/search-workflow.sh
# Print search results
cat examples-tmp/search-workflow/drugbank-all-q1-results.txt
cat examples-tmp/search-workflow/drugbank-all-q30-mostsimilars-results.txt
Final search results for searching norbornane (SMILES: C1CC2CCC1C2) against the drugbank-all dataset (from file examples-tmp/search-workflow/drugbank-all-q1-results.txt):
Query Target Dissimilarity
0 Camphane 0.08333333333333333
First few lines of the final search results for searching the 5 most similars for members of the vitamins dataset against the drugbank-all dataset (from file examples-tmp/search-workflow/drugbank-all-q30-mostsimilars-results.txt):
Query Target Dissimilarity
0 Vitamin A 0.0
0 Alitretinoin 0.14814814814814814
0 Tretinoin 0.14814814814814814
0 Isotretinoin 0.14814814814814814
0 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene 0.1956521739130435
1 Alitretinoin 0.14814814814814814
1 Tretinoin 0.14814814814814814
1 Isotretinoin 0.14814814814814814
1 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene 0.1956521739130435
1 4-Oxoretinol 0.23333333333333334
2 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene 0.02631578947368421
2 Vitamin A 0.17391304347826086
2 (6e)-6-[(2e,4e,6e)-3,7-Dimethylnona-2,4,6,8-Tetraenylidene]-1,5,5-Trimethylcyclohexene 0.2
2 Alitretinoin 0.2962962962962963
2 Tretinoin 0.2962962962962963
3 Thiamine 0.0
3 Thiamin Phosphate 0.15757575757575756
....
Launch Web UI
Launch script examples/rest-api-small.sh. This script calculates CFP-7 fingerprints for the nci-250k dataset and launches a web based user interface where real time similarity search of the dataset is available. Profiling and execution statistics data is also collected and exposed. For details on launching the web ui and on other self contained examples see document REST API example.
examples/rest-api-small.sh -b
Preprocessing time is expected to be under a minute on an average machine. After the preprocessing the scripts starts the web ui server which listens http://localhost:8085. When option -b passed to the example script the Web UI will try to launch the default web browser.
....
Try to launch browser on http://localhost:8085/index.html
A browser window is opened and an index page is displayed:
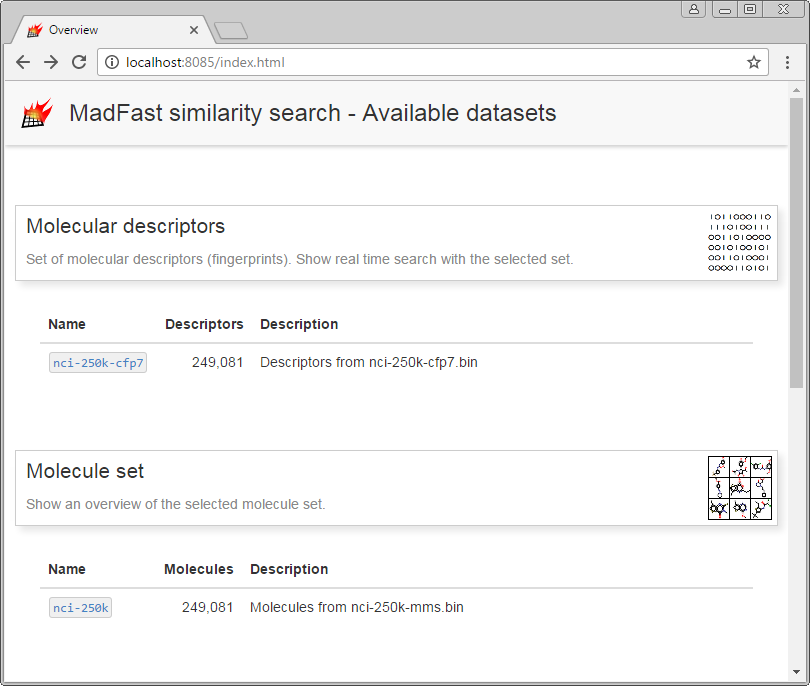
On the displayed index page select nci-250k-cfp7 from Molecular descriptors to launch the real time similarity search example.

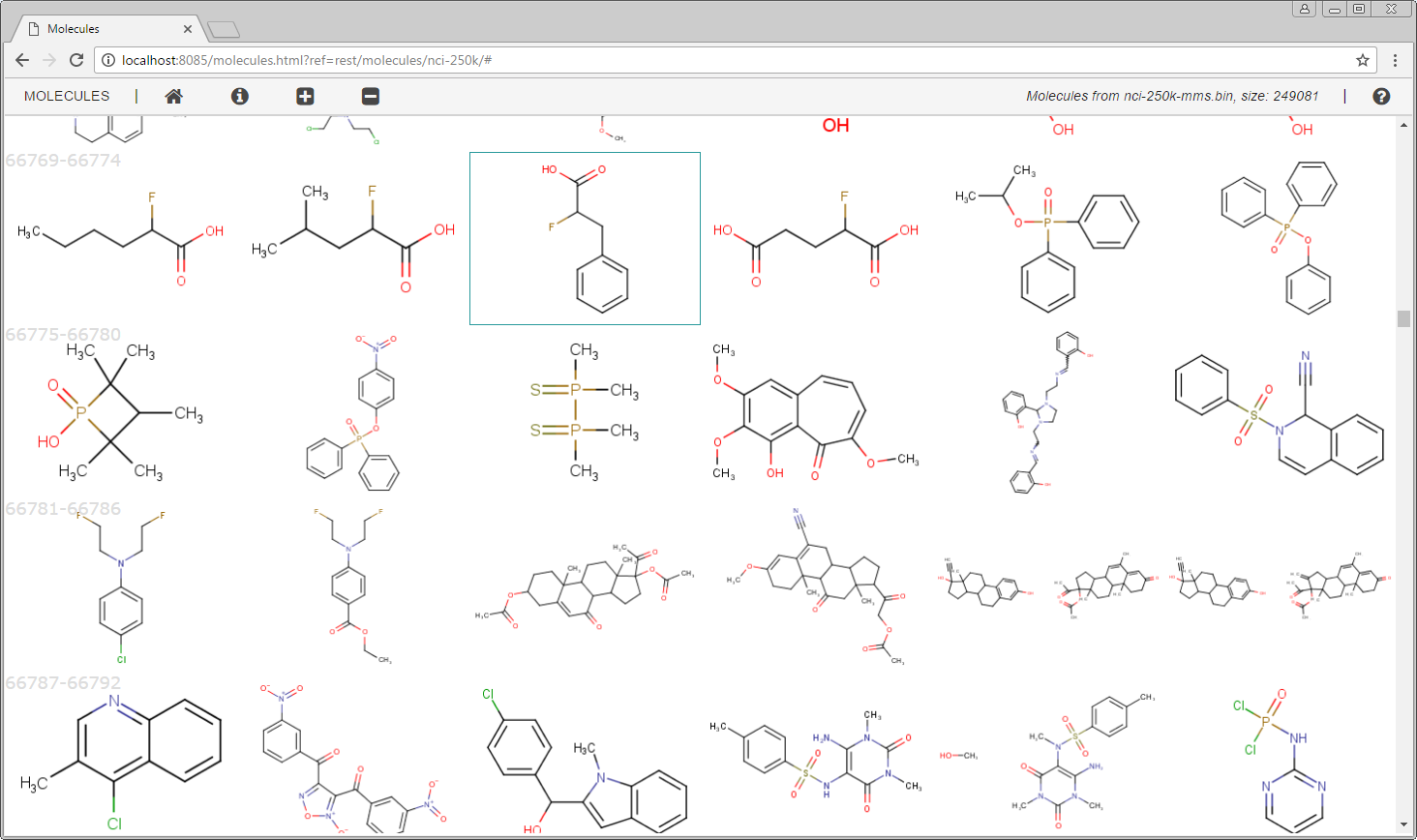
Or select nci-250k from Molecule set to see the structures in the set.
Scripts exposing multiple, larger datasets, multiple descriptors are also available (rest-api-XXXX.sh). For an overview see document Self contained examples.